Ontologies
last updated at 2023-02-28The term ontology has its origin in the philosophic question about the very nature of our existence. Here, we do not want to engage into a discussion that could fill whole books.
In our context, an ontology comprises a conceptualized collection of well-defined terms and their relationships for a specific research domain, e.g., plant sciences. Each term and relationship consists of a unique ID, a label and a definition that are curated manually by experts of the respective domain. Therefore, it can be used to semantically annotate and describe research data in a way that makes it both human- and machine-readable. Using the human-readable definition each researcher can understand and correctly reuse specific terms, while a computer can use the unique ID to reference any term from the ontology. If one term will be replaced by a more accurate term it will not be deleted from the ontology but rather marked as "obsolete". This ensures that older documents that reference the old ontology term remain valid. Using the relationships within the ontology, experimental data can be structured and easily compared to other structured datasets and infer new information.
All knowledge within an ontology is described through axiom triples in a subject-predicate-object syntax. An example for this is the following triplet:
The "phyllome" (subject) is "part of" (predicate) the "shoot system" (object).
There are several terms (depending on the ontology format) that are used to describe specific parts of an ontology:
| OWL term | OBO term | Explanation | OWL Manchester Example |
|---|---|---|---|
| Class | Term (stanza) | Define groups of individuals that belong together because they share some properties. | Class: PO_0006001 Annotations: rdfs:label "phyllome" |
| ObjectProperty | Typedef (stanza) | Relations between instances of two classes. | ObjectProperty: BFO_0000050 Annotations: rdfs:label "part of" |
| DataProperty | no equivalent | Relations between instances of classes and datatypes. | DataProperty: ARC_00000243 Annotations: rdfs:label "email" |
| AnnotationProperty | property_value (tag) | Define that a property is an annotation. | AnnotationProperty: dc:creator |
| Individual | Instance (stanza) | Instances of classes that can be related to other instances through properties. | Individual: ARC_00000500 Annotations: rdfs:label "A. thaliana" |
| Domain | domain (tag) | Subject of a relationship triplet. | ObjectProperty: BFO_0000050 Annotations: rdfs:label "part of" Domain: PO_0006001 |
| Range | range (tag) | Object of a relationship triplet. | ObjectProperty: BFO_0000050 Annotations: rdfs:label "part of" Range: PO_0009006 |
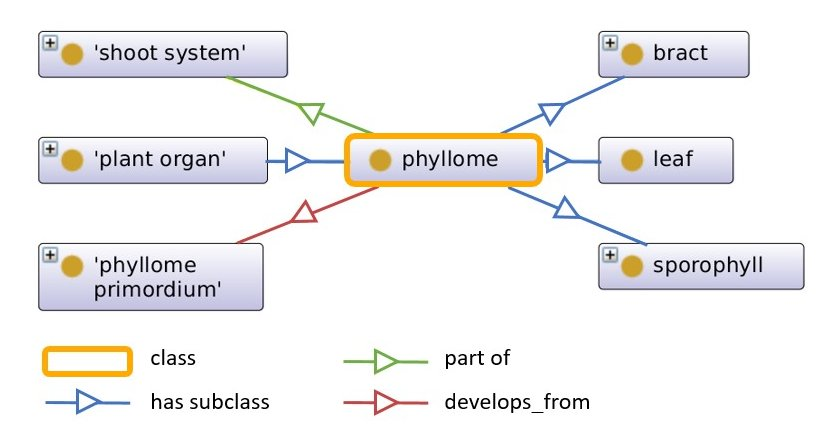
Fig. 1: Excerpt of the plant ontology. Boxes describe classes, arrows describe object properties. The class “phyllome” is defined as a subclass of “plant organ”. The class “phyllome” has three subclasses: “bract”, “leaf” and “sporophyll”. It is furthermore defined as “part of” the class “shoot system” and the "phyllome" “develops_from” the “phyllome primordium”.
Ontologies can be used for many different purposes. As such, there are also multiple languages which an ontology can be written in, as well as many formats. Some of the most important relevant for our context include the Web Ontology Language (OWL) and Open Biomedical Ontologies (OBO) format. OBO was mainly used by the biomedical community (see section OBO Foundry) before the rise of OWL and today is considered a dialect of OWL.
The OBO format is the simplest format and not only machine-readable but also easily read by humans. As such, it is easy to edit and review, but also lacks multiple features only available in other formats. For instance, it is not possible to define DatatypeProperties in OBO, which are used in OWL ontologies.
OBO example:
When it comes to OWL ontologies, there are several common formats: RDF/XML, OWL/XML, OWL Functional Syntax and OWL Manchester Syntax. All formats are more complex than the easy-to-read OBO format. While RDF/XML is the best format for providing a suitable input format file for a multitude of tools, it is very complex and more difficult to review without a good understanding of the format. Review and editing can be more easily performed using the OWL Manchester Syntax, which provides all information about a concept and its usages within the ontology in one paragraph.
RDF/XML example:
OWL Manchester example:
Since the OBO format does not cover all aspects that can be included in OWL formatted ontologies, conversion between the formats can be problematic. Generally, it is possible to convert OBO files into OWL files using conversion tools such as the OWL API, since OBO can be interpreted as a reduced OWL form. However, when trying to convert OWL files to OBO files, there can be significant information loss when concepts foreign to OBO (such as DatatypeProperties mentioned above) are used.
The Open Biological Biomedical Ontologies (OBO) Foundry is a team of scientists interested in the development, collection and maintenance of ontologies for the biological and biomedical domain of life sciences.
The OBO Foundry is open for contribution from everyone. For an ontology to be accepted into the OBO Foundry, it must follow the OBO Foundry Principles:
It must be open and it needs to be licensed (e.g. CC-BY 3.0).
A common format must be used (see section OBO and OWL format).
Each term should have a unique identifier by using a unique prefix (such as GO) followed by a numerical ID, thereby resulting in an unique OBO Foundry URL.
Every new version of an ontology must be versioned using documented procedures.
The domain or matter of the ontology must be described.
Each term in an ontology must be described in a human-readable form.
Relations between different items of the ontology should be described using defined vocabulary described by the Relations Ontology (RO).
Documentations for the ontology must be available. Here, the best way is to provide a GitHub repository.
It should be documented that multiple independent people or organizations use the ontology.
The ontology should be open to contribution from anyone.
One person should be listed as contact and responsible person for the ontology.
The naming of for elements such as classes, subclasses etc. should follow naming conventions.
- The ontology needs to be maintained.
- Ontology developers MUST offer channels for community participation and SHOULD be responsive to requests.